The Niu research group is multidisciplinary, combining biochemistry, electrophysiology, molecular biology, molecular folding/docking, and neuroscience to study significant problems in human health. A major focus of our current study is on glutamate ion channel proteins. These channel proteins are indispensable to brain activities such as learning and memory, and malfunction of these proteins has been implicated in neurological diseases such as ALS, stroke, epilepsy, and neuropathic pain. We are working on project areas with broader impact potential, such as structure-function relationship of glutamate ion channels and drug discovery, by developing RNA aptamers and small-molecule regulatory agents (i.e., both positive and negative regulators) as potential drug candidates. RNA hydrogel becomes one of the projects stemmed from our discovery that an RNA aptamer discovered in our group is capable of self-assembling into RNA hydrogel. We are also testing highly potent and highly selective RNA aptamers we have discovered with animal models of human diseases, e.g., amyotrophic lateral sclerosis (ALS) and spinal cord induced neuropathic pain, through tissue and behavioral studies.
About
Research
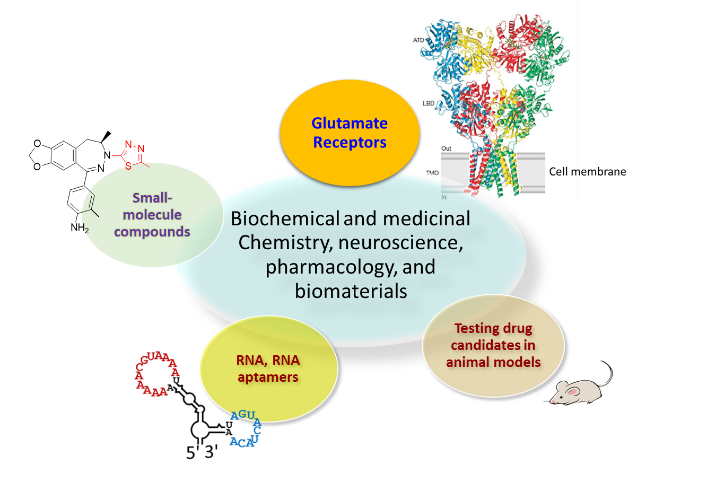

Structure-function relationship of glutamate ion channels
The three-dimensional structure of a protein defines not only its size and shape but also its function. Understanding the relationship between protein structure and function is one of the foremost challenges in finding ways to regulate the in vivo function of a protein with precision. Projects in this area of our lab are focused on rapid kinetic measurement of the channel-opening rate on the microsecond time scale by using a laser-pulse photolysis technique with “cage glutamate.” These measurements are essential towards a better understanding of the mechanism of action for various subunits/isoforms of glutamate ion channels and developing effective means and molecular reagents to precisely control the function of various isoforms and subunits of these glutamate ion channel receptors. Currently, we are focusing on AMPA and kainate receptor subtypes and their isoforms in each subtype.
Developing small-molecule, regulatory agents (inhibitors and potentiators) as potential drug candidates
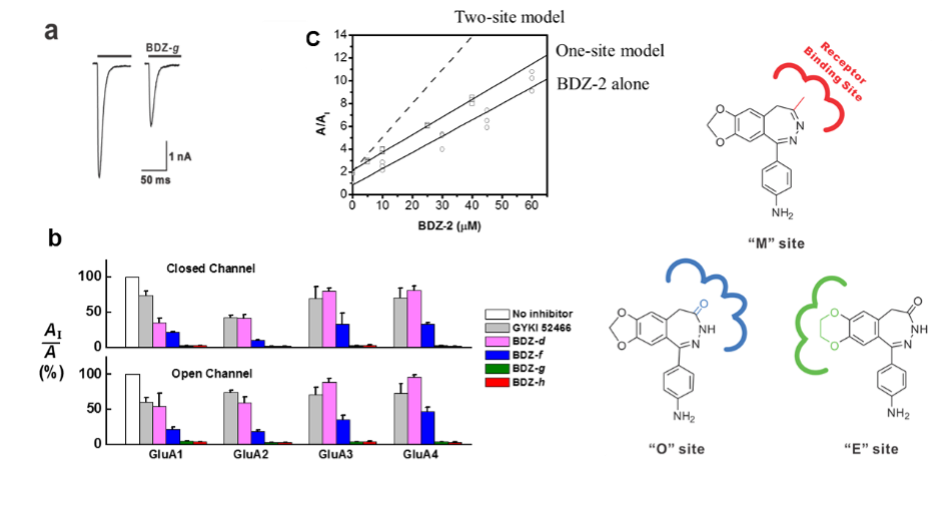
This area focuses on developing small-molecule compounds and understanding the mechanism of action as well as the site of interaction with receptor targets. The goal is to modulate the activity of the target in vivo with high potency and selectivity. These small-molecule compounds, prepared by synthetic chemistry, are potential drug candidates for a number of neurological diseases such as amyotrophic lateral sclerosis (ALS), epilepsy and stroke. Positive modulators are potential drug candidates for cognitive deficiency such as memory deficits in Parkinson’s disease. We also characterize the structure-activity relationship for these compounds.
RNA: Aptamers as potential drug candidates
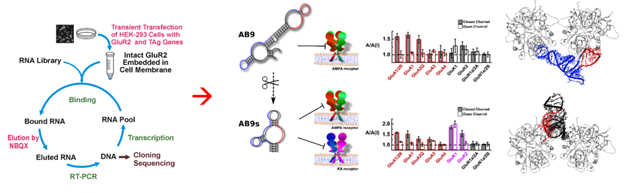
One of the core components in our research is the discovery of RNA molecules as water-soluble, potent and selective antagonists or positive modulators (potentiators) of glutamate ion channels. We use SELEX technique to isolate these useful aptamers from a large RNA library (about 1,014 sequences). We also make chemically modified RNA aptamers by replacing 2’-OH group in ribose so these RNA aptamers are long lasting and are therefore amenable for in vivo use as drug candidates for CNS diseases.
RNA: Folding and function
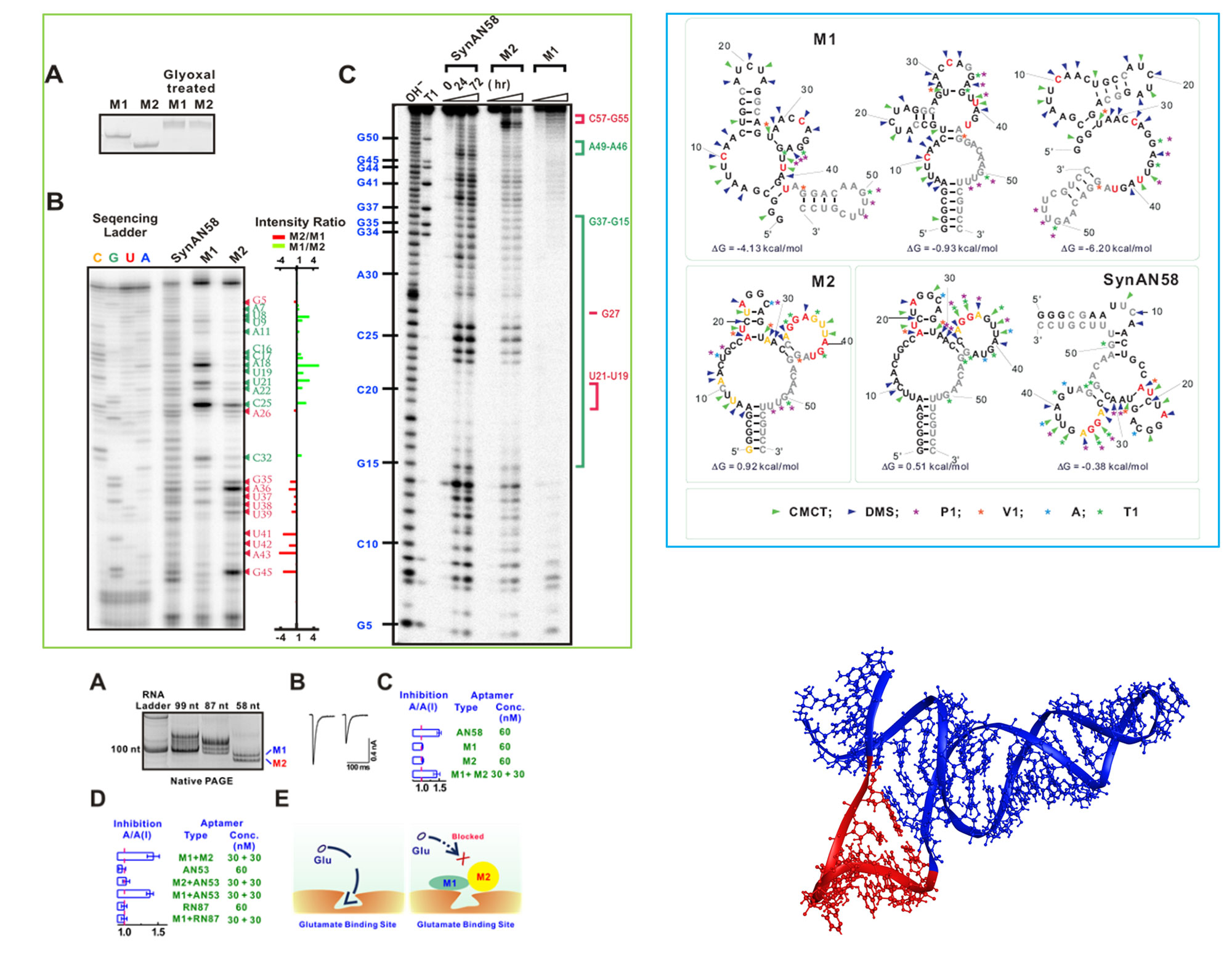
This area of research focuses on understanding of RNA folding into stable structures for a myriad of functions, such as modulating protein activities as drug candidates. RNA has extensive intramolecular interactions that cause it to fold from a linear molecule with specific sequence into an array of complex structures. RNA folding begins as the transcript is synthesized by RNA polymerase. The formation of RNA structures, such as hairpins, on the nascent transcript can create physical barriers that prevent backtracking and promote certain pathways favor specific tertiary structures with unique functional properties. Here we study the folding of an RNA, which can adopt multiple structures (not reversible, dynamic conformations) with distinct functions from transcription (i.e., copying of a single DNA sequence into RNAs by RNA polymerase).
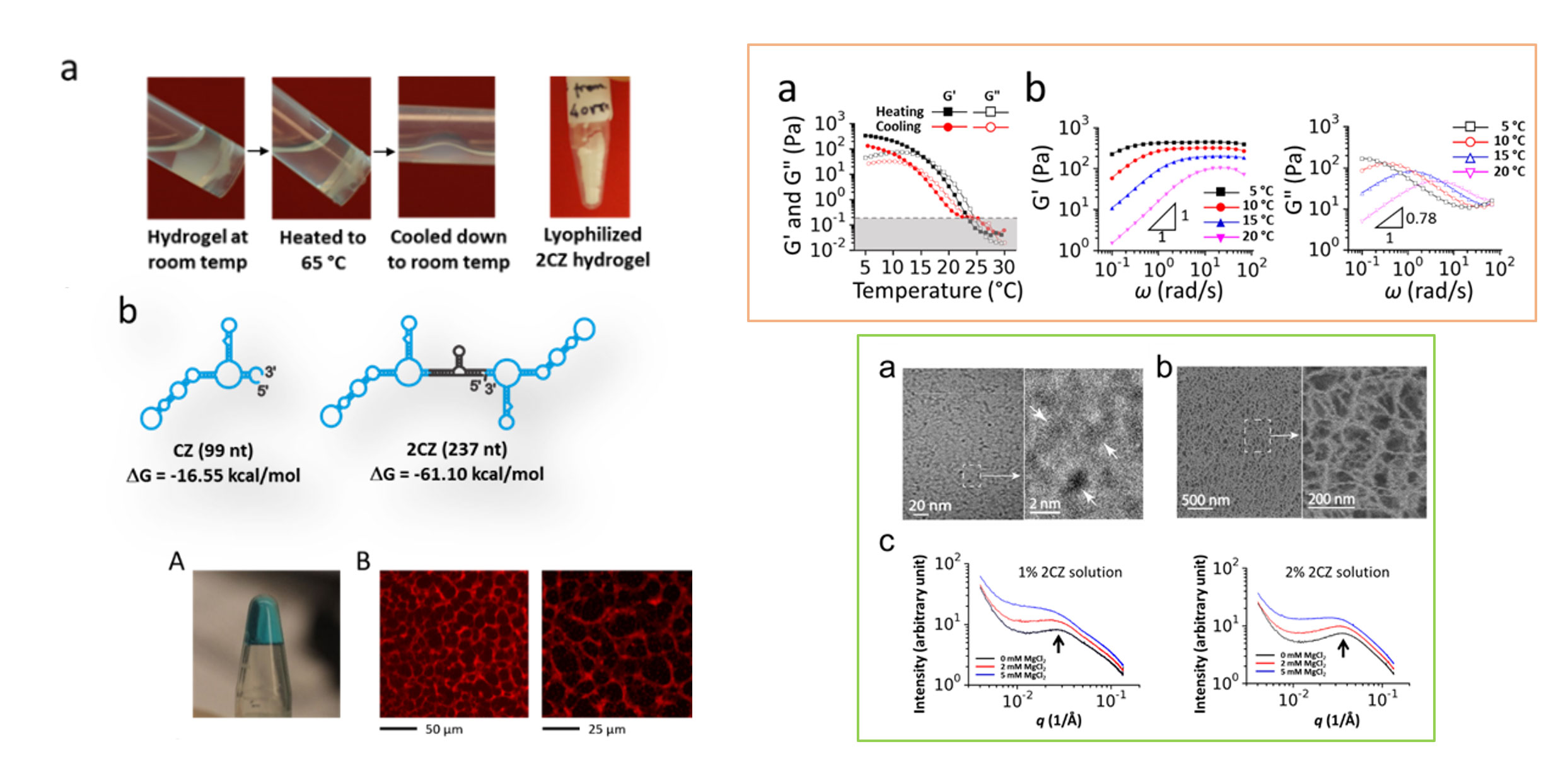
RNA: Hydrogel and biomaterials
Hydrogels are supramolecular assemblies with both solute transport properties like liquids and mechanical properties like elastomers. To date, every type of biomolecule — except RNA — is capable of forming hydrogel. Recently, we have found a unique RNA that can self-assemble, without any cross-linker or any external support matrix, to form hydrogel. In this case, a single RNA is the monomer, which contains proper recognition domains for intermolecular, non-covalent interactions in order to self-assemble into the hydrogel network structure. Hydrogels formed from biomolecules like RNA are generally biocompatible and can be chemically and biologically decorated and functionalized, making them particularly useful for tissue engineering and drug delivery, etc. We are identifying design modules and making various RNA hydrogels with different mechanical and physical properties. We are testing them using a variety of techniques such as SAXS, cryo-SEM and cryo-TEM.
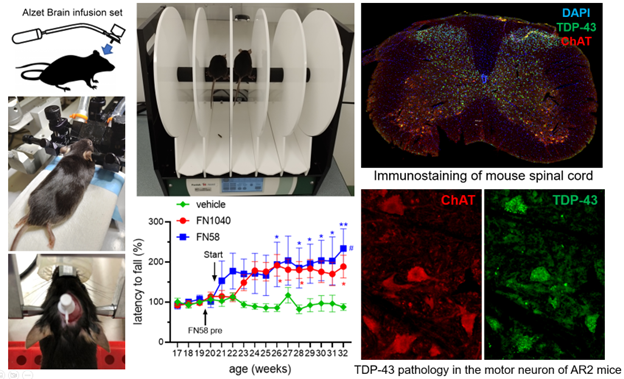
Testing RNA aptamers with animal models of human diseases
Discovery of a drug candidate in terms of its efficacy and its potential toxicological effects requires in vivo study, and animal models are the most practical tools for in vivo, pre-clinical study of a drug candidate before its application in clinical trials. Currently, we are testing a group of RNA aptamers with two transgenic mouse models that mimic familial and sporadic ALS. ALS is a fatal type of motor neuron disease, and currently there is no cure. In one of our studies, for example, we examined the efficacy of an aptamer infused through intraventricular injection on mice with dysregulated AMPA receptor activity. We found that administration of the aptamer rescued both the number and the size of the motor neurons, and the mice performed better in motor function. We are also testing our RNA aptamers with a rat model that mimics neuropathic pain induced by spinal cord injury (SCI). Neuropathic pain caused by a lesion or disease of the somatosensory nervous system is a common chronic pain condition with major impact on quality of life. Most of the available treatments for neuropathic pain have moderate efficacy and present side effects that limit their use. Our preliminary study has shown that administration of RNA aptamers to rats with SCI has generated strong, long-lasting analgesic efficacy without any detectable side effects.
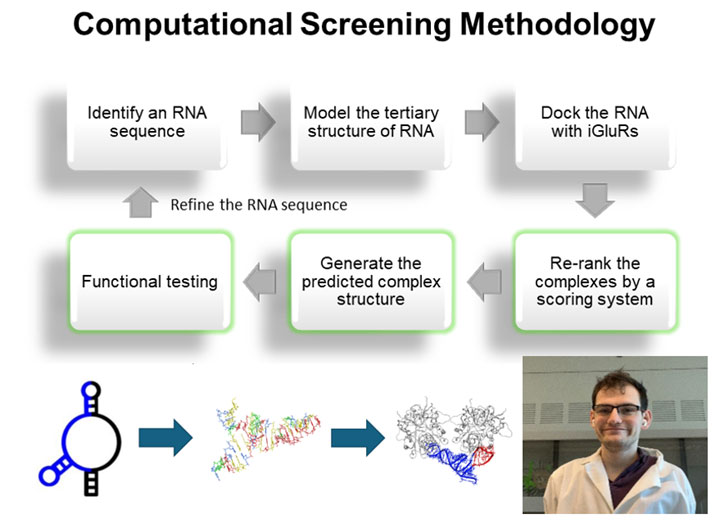
RNA-receptor molecular docking: Using computational approach for discovery of highly selective RNA aptamers
Computational modeling of RNA-glutamate receptor interactions has been a recent addition as a tool for our effort to develop RNA aptamers with high potency and selectivity. This method is especially useful when combined with RNA sequence mutations and then functional testing of the sequence variants with individual glutamate receptor channels using electrophysiology. The diagram below shows a flowchart of our approach.
Publications
- Saunders, N. & Niu, L. (2025) A comparative study of NETO1 and NETO2 on channel opening kinetics of GluK2 kainate receptors. Journal of Biological Chemistry. Published online, Oct 4, 2025
- Jergova, S., Huang, Z., Lin, C.Y., Akamatsu, M., Sagen, J. & Niu, L. (2025) Isolation, characterization and testing of RNA aptamers targeting glutamate receptors in a rat spinal cord injury pain model. Communications Biology Sep 26;8(1):1375. doi: 10.1038/s42003-025-08772-8.
- Ingenito, S.R., Saunders, N., Lininger, K.J. & Niu, L. (2025) GluK2 kainate receptor subunit-selective, potentiating RNA aptamer. Sci. Reports 15:32405.
- Akamatsu, M., Yamashita, T., Teramoto, S., Huang, Z., Lynch, J., Toda, T., Niu, L., & Kwak, S. (2022) Testing of the therapeutic efficacy and safety of AMPA receptor RNA aptamers in an ALS mouse model. Life Science Alliance, 5(4).
- Huang, Z., & Niu, L. (2021) RNA aptamers for AMPA receptors. Neuropharmacology, 199, 108761.
- Jaremko, W., Huang, Z., Karl, N., Pierce, V. D., Lynch, J., & Niu, L. (2020) A kainate receptor–selective RNA aptamer. Journal of Biological Chemistry, 295(19), 6280–6288.
- Huang, Z., & Niu, L. (2019) Developing RNA aptamers for potential treatment of neurological diseases. Future Medicinal Chemistry, 11(6), 551–565.
- Jaremko, W. J., Huang, Z., Wen, W., Wu, A., Karl, N., and Niu, L. (2017) Identification and characterization of RNA aptamers: A long aptamer blocks the AMPA receptor and a short aptamer blocks both AMPA and kainate receptors. J Biol Chem 292, 7338-7347.
- Huang, Z., Kangovi, G. N., Wen, W., Lee, S., and Niu, L. (2017) An RNA Aptamer Capable of Forming Hydrogel by Self-Assembly. Biomacromolecules. 18, 2056-2063.
- Han, Y., Lin, C. Y., and Niu, L. (2017) Functional Roles of the Edited Isoform of GluA2 in GluA2-Containing AMPA Receptor Channels. Biomacromolecules. 18, 2056-2063.
- Espahbodinia, M., Ettari, R., Wen, W., Wu, A., Shen, Y. C., Niu, L., Grasso, S., and Zappala, M. (2017) Development of novel N-3-bromoisoxazolin-5-yl substituted 2,3-benzodiazepines as noncompetitive AMPAR antagonists. Bioorg Med Chem 25, 3631-3637.
- Wang, C., Wu, A., Shen, Y. C., Ettari, R., Grasso, S., and Niu, L. (2015) Mechanism and site of inhibition of AMPA receptors: substitution of one and two methyl groups at the 4-aminophenyl ring of 2,3-benzodiazepine and implications in the "E" site. ACS Chem Neurosci 6, 1371-1378.
- Lin, C. Y., Huang, Z., Wen, W., Wu, A., Wang, C., and Niu, L. (2015) Enhancing Protein Expression in HEK-293 Cells by Lowering Culture Temperature. PloS one 10, e0123562.
- Huang, Z., Lin, C. Y., Jaremko, W., and Niu, L. (2015) HPLC purification of RNA aptamers up to 59 nucleotides with single-nucleotide resolution. Methods in molecular biology (Clifton, N.J.) 1297, 83-93.
- Wu, A., Wang, C., and Niu, L. (2014) Mechanism of inhibition of the GluA1 AMPA receptor channel opening by the 2,3-benzodiazepine compound GYKI 52466 and a N-methyl-carbamoyl derivative. Biochemistry 53, 3033-3041.
- Wang, C., Han, Y., Wu, A., Solyom, S., and Niu, L. (2014) Mechanism and site of inhibition of AMPA receptors: pairing a thiadiazole with a 2,3-benzodiazepine scaffold. ACS Chem Neurosci 5, 138-147.
- Wang, C., and Niu, L. (2013) Mechanism of inhibition of the GluA2 AMPA receptor channel opening by talampanel and its enantiomer: the stereochemistry of the 4-methyl group on the diazepine ring of 2,3-benzodiazepine derivatives. ACS Chem Neurosci 4, 635-644.
- Huang, Z., Jayaseelan, S., Hebert, J., Seo, H., and Niu, L. (2013) Single-nucleotide resolution of RNAs up to 59 nucleotides by high-performance liquid chromatography. Analytical biochemistry 435, 35-43.
- Han, Y., Wang, C., Park, J. S., and Niu, L. (2012) Channel-opening kinetic mechanism of wild-type GluK1 kainate receptors and a C-terminal mutant. Biochemistry 51, 761-768.
- Wang, Z., Zhang, J., Chen, P., Zhou, X., Yang, Y., Wu, S., Niu, L., Han, Y., Wang, L., Chen, P., Boey, F., Zhang, Q., Liedberg, B., and Zhang, H. (2011) Label-free, electrochemical detection of methicillin-resistant Staphylococcus aureus DNA with reduced graphene oxide-modified electrodes. Biosens Bioelectron 26, 3881-3886.
- Wang, C., Sheng, Z., and Niu, L. (2011) Mechanism of inhibition of the GluA2 AMPA receptor channel opening: consequences of adding an N-3 methylcarbamoyl group to the diazepine ring of 2,3-benzodiazepine derivatives. Biochemistry 50, 7284-7293.
- Ritz, M., Wang, C., Micale, N., Ettari, R., and Niu, L. (2011) Mechanism of Inhibition of the GluA2 AMPA Receptor Channel Opening: the Role of 4-Methyl versus 4-Carbonyl Group on the Diazepine Ring of 2,3-Benzodiazepine Derivatives. ACS Chem Neurosci 2, 506-513.
- Park, J. S., Wang, C., Han, Y., Huang, Z., and Niu, L. (2011) Potent and selective inhibition of a single alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor subunit by an RNA aptamer. J Biol Chem 286, 15608-15617.
- Huang, Z., Han, Y., Wang, C., and Niu, L. (2010) Potent and selective inhibition of the open-channel conformation of AMPA receptors by an RNA aptamer. Biochemistry 49, 5790-5798.
- Pei, W., Huang, Z., Wang, C., Han, Y., Park, J. S., and Niu, L. (2009) Flip and flop: a molecular determinant for AMPA receptor channel opening. Biochemistry 48, 3767-3777.
- Huang, Z., Pei, W., Han, Y., Jayaseelan, S., Shekhtman, A., Shi, H., and Niu, L. (2009) One RNA aptamer sequence, two structures: a collaborating pair that inhibits AMPA receptors. Nucleic acids research 37, 4022-4032.
- Pei, W., Ritz, M., McCarthy, M., Huang, Z., and Niu, L. (2007) Receptor occupancy and channel-opening kinetics: a study of GLUR1 L497Y AMPA receptor. J Biol Chem 282, 22731-22736.
- Pei, W., Huang, Z., and Niu, L. (2007) GluR3 flip and flop: differences in channel opening kinetics. Biochemistry 46, 2027-2036. J Biol Chem 282, 22731-22736.
- Kornreich, B. G., Niu, L., Roberson, M. S., and Oswald, R. E. (2007) Identification of C-terminal domain residues involved in protein kinase A-mediated potentiation of kainate receptor subtype 6. Neuroscience 146, 1158-1168.
- Huang, Z., Pei, W., Jayaseelan, S., Shi, H., and Niu, L. (2007) RNA aptamers selected against the GluR2 glutamate receptor channel. Biochemistry 46, 12648-12655.
- Li, G., Sheng, Z., Huang, Z., and Niu, L. (2005) Kinetic mechanism of channel opening of the GluRDflip AMPA receptor. Biochemistry 44, 5835-5841.
- Huang, Z., Li, G., Pei, W., Sosa, L. A., and Niu, L. (2005) Enhancing protein expression in single HEK 293 cells. Journal of neuroscience methods 142, 159-166.
- Li, G., and Niu, L. (2004) How fast does the GluR1Qflip channel open?. J Biol Chem 279, 3990-3997.
- Li, G., Pei, W., and Niu, L. (2003) Channel-opening kinetics of GluR2Q(flip) AMPA receptor: a laser-pulse photolysis study. Biochemistry 42, 12358-12366.
- Li, G., Oswald, R. E., and Niu, L. (2003) Channel-opening kinetics of GluR6 kainate receptor. Biochemistry 42, 12367-12375.
- Niu, L., Kim, J. M., and Khorana, H. G. (2002) Structure and function in rhodopsin: asymmetric reconstitution of rhodopsin in liposomes. Proc Natl Acad Sci U S A 99, 13409-13412.
- Hess, G. P., Ulrich, H., Breitinger, H. G., Niu, L., Gameiro, A. M., Grewer, C., Srivastava, S., Ippolito, J. E., Lee, S. M., Jayaraman, V., and Coombs, S. E. (2000) Mechanism-based discovery of ligands that counteract inhibition of the nicotinic acetylcholine receptor by cocaine and MK-801. Proc Natl Acad Sci U S A 97, 13895-13900.
- Bruel, C., Cha, K., Niu, L., Reeves, P. J., and Khorana, H. G. (2000) Rhodopsin kinase: two mAbs binding near the carboxyl terminus cause time-dependent inactivation. Proc Natl Acad Sci U S A 97, 3010-3015.
- Gee, K. R., Niu, L., Schaper, K., Jayaraman, V., and Hess, G. P. (1999) Synthesis and photochemistry of a photolabile precursor of N-methyl-D-aspartate (NMDA) that is photolyzed in the microsecond time region and is suitable for chemical kinetic investigations of the NMDA receptor. Biochemistry 38, 3140-3147.
- Niu, L., Wieboldt, R., Ramesh, D., Carpenter, B. K., and Hess, G. P. (1996) Synthesis and characterization of a caged receptor ligand suitable for chemical kinetic investigations of the glycine receptor in the 3-microseconds time domain. Biochemistry 35, 8136-8142.
- Niu, L., Vazquez, R. W., Nagel, G., Friedrich, T., Bamberg, E., Oswald, R. E., and Hess, G. P. (1996) Rapid chemical kinetic techniques for investigations of neurotransmitter receptors expressed in Xenopus oocytes. Proc Natl Acad Sci U S A 93, 12964-12968.
- Niu, L., Gee, K. R., Schaper, K., and Hess, G. P. (1996) Synthesis and photochemical properties of a kainate precursor and activation of kainate and AMPA receptor channels on a microsecond time scale. Biochemistry 35, 2030-2036.
- Niu, L., Abood, L. G., and Hess, G. P. (1995) Cocaine: mechanism of inhibition of a muscle acetylcholine receptor studied by a laser-pulse photolysis technique. Proc Natl Acad Sci U S A 92, 12008-12012.
- Hess, G. P., Niu, L., and Wieboldt, R. (1995) Determination of the chemical mechanism of neurotransmitter receptor-mediated reactions by rapid chemical kinetic methods. Ann N Y Acad Sci 757, 23-39.
- Wieboldt, R., Gee, K. R., Niu, L., Ramesh, D., Carpenter, B. K., and Hess, G. P. (1994) Photolabile precursors of glutamate: synthesis, photochemical properties, and activation of glutamate receptors on a microsecond time scale. Proc Natl Acad Sci U S A 91, 8752-8756.
- Ramesh, D., Wieboldt, R., Niu, L., Carpenter, B. K., and Hess, G. P. (1993) Photolysis of a protecting group for the carboxyl function of neurotransmitters within 3 microseconds and with product quantum yield of 0.2. Proc Natl Acad Sci U S A 90, 11074-11078.
- Niu, L., and Hess, G. P. (1993) An acetylcholine receptor regulatory site in BC3H1 cells: characterized by laser-pulse photolysis in the microsecond-to-millisecond time region. Biochemistry 32, 3831-3835.
Members
Principal Investigator

Li Niu
Professor of Chemistry
LSRB 1060
[email protected]
Lab Members

Anna Liu
MS Student

Leyla Caliskan
Undergraduate Student

Jessica Rosales
Undergraduate Student

Nicole Aragona
Undergraduate Student
Alumni
Rodica Doaga, PhD
Yan Han, PhD
Zhen Huang, PhD
Samantha Ingenito, PhD
William Jaremko, PhD
Sabari Jayaseelan, PhD
Gang Li, PhD
Janet Lynch, PhD
Jae Seon Park, PhD
Weimin Pei, PhD
Mohammad Qneibi, PhD
Zhenyu Sheng, PhD
Congzhou Wang, PhD
Wei Wen, PhD
Andrew Wu, PhD
Tani Yumeki, PhD
Jeff Hebert, MS
Nicholas Karl, MS
Chi-Yen Lin, MS
Vincen Pierce, MS
Mark Ritz, MS
Hyojung Seo, MS
Yuchuan Shen, MS
Deon Teh, MS
Contact
1400 Washington Avenue
Albany, NY 12222
United States
Photos