Dr. Alan Chen presents R2D2- a new method for investigating RNA folding pathways
(January 15th, 2021) Dr. Alan Chen, Associate Professor of Chemistry at the University at Albany and RNA Institute Faculty member, has teamed up with researchers at Ohio State and Northwestern University to develop a new method to better understand cotranscriptional RNA folding.
Each time an RNA is made in the cell, it will begin to fold even before the entire sequence is finished being made. This “cotranscriptional folding” process helps steer the nascent RNA to rapidly adopt the specific conformation required for biological activity.
Despite this biological importance, the actual folding pathways that RNAs undergo during transcription are still not well understood, limiting our knowledge of the most basic aspects of RNA biology.
Efforts to predict cotranscriptional RNA folding pathways are in early stages. Although the RNA genetic sequences and the 3D conformations they ultimately adopt can be experimentally measured, there is no experiment capable of directly detecting what an RNA folding pathway looks like in atomistic detail.
The research team developed a new method called reconstructing RNA dynamics from data (R2D2) to uncover details of cotranscriptional folding. This method combines nucleotide-resolution experimental RNA structure chemical probing with computational algorithms to discern base-pairing patterns from nascent RNAs of varying transcript length data (both developed by Julius Lucks’ team at Northwestern University). These patterns were then used to “steer” atomistic computer simulations carried out by the UAlbany team led by Dr. Chen along with postdoc Paul Gasper and undergraduate researcher Simi Kaur. These highly complex molecular dynamics simulations of over 300,000 atoms required hundreds of thousands of CPU-hours on the COMET supercomputer at UC San Diego courtesy of the NSF (which funded Dr. Chen’s research).
The combination of rapid chemical probing, bioinformatic analysis, and molecular simulations resulted in “molecular movies” that reconstructed the RNA folding pathway in atomistic detail.
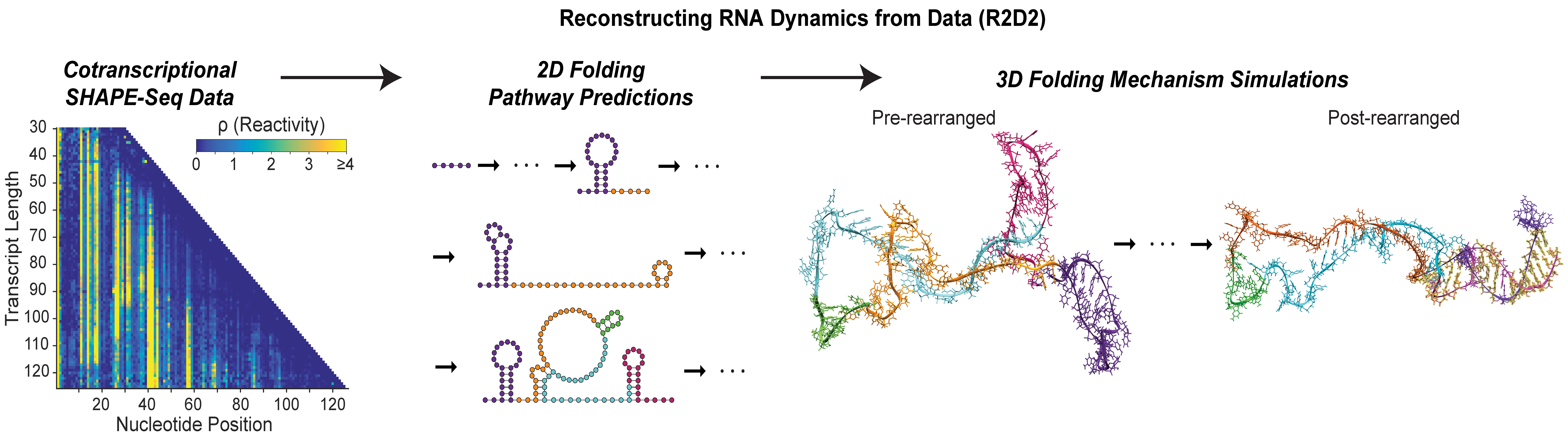
Figure 1: Rapid chemical probing experiments on different transcript lengths (left) are used to reconstruct base-pairing patterns (center) used to constrain molecular simulations of the folding pathway (right)
When applied to E. coli’s signal recognition particle RNA, this study both confirmed the existence of previously hypothesized folding intermediates and provided new evidence that the observed rapid conformational switching was the result of a “toehold” strand-displacement mechanism.
“This was really exciting because toehold displacement based conformational switching is a design strategy ‘invented’ by DNA nanotechnologists but never before documented in a naturally occurring system” explained Dr. Chen. “It turns out, nature already discovered it hundreds of millions of years ago. Undoubtedly, there are countless more design principles like this buried in our genomes, we just need to know where to look”.
Understanding how evolution has optimized RNA sequences to rapidly fold into specific shapes without making mistakes will enable the rational engineering of RNAs for biotechnology applications such as genetic engineering and biosensing.
Read the full publication here
Ground-breaking films show RNA's complex curves take shape - Nature
RNA ties itself in knots, then unties itself in mesmerizing video - LiveScience
New videos show RNA as it’s never been seen - Science Magazine


