Studying the Shape of Gum Disease: $2.3 Million Grant Aids Research into New Bacterial Target for Dental Treatments
By Erin Frick
ALBANY, N.Y. (Sept. 1, 2022) — Our mouths play host to a rich community of microorganisms. Some perform essential functions, such as those that aide digestion. Others are pathogenic and cause disease.
Scientists in the College of Arts and Sciences' departments of biology and mathematics are using RNA sequencing to study the role of dental plaque in the progression of periodontal disease. Their work is being funded by a $2.3 million grant from the National Institute of Dental and Craniofacial Research, an arm of the National Institutes of Health.
Every day, we raise paste-laden toothbrushes to fight plaque — the biofilm that makes teeth feel fuzzy and harms gum health. Understanding how the structure of plaque influences the ability of harmful bacteria to take hold can inform new treatments for periodontal disease, which is a major contributor to health disparity along socioeconomic lines worldwide.
“Common methods of periodontal care, like brushing and mouthwash, target plaque broadly,” Alex Valm, principal investigator on the project and assistant professor of biology working in the RNA Institute, explains. “Like a broad-spectrum antibiotic, this approach wipes out good bacteria along with the bad. Yet, even people who have access to basic hygienic resources experience gum disease. So what are we missing? Here we are asking — is there an organism in plaque playing an unknown role in gum disease? If so, it could become an important target for new therapeutics.”
The research team will use RNA sequencing and imaging to study microbial species present in dental plaque and track changes in the microbial community as gum disease progresses.
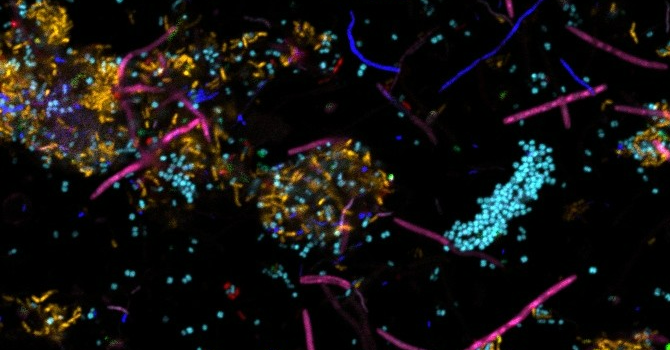
“Imaging plaque allows us to identify which microbial species make up the biofilm and where they exist in the structure,” Valm says. “However, the location of the species does not tell the whole story. We will also examine how microbial gene expression changes through periodontal disease progression — as the dental plaque biofilm develops from one that promotes health to one that induces inflammatory disease. This is what the RNA sequencing will tell us. We need to know not just who’s there and where; we also need to know what they are doing.”
Plaque structure is a central mechanism affecting disease risk. The microbial community that makes up dental plaque includes long filamentous organisms that can protrude from tens up to hundreds of microns from the tooth enamel or root surface. When a pathogen encounters one of these protrusions, this incites an inflammatory response — aka, the start of gum disease.
Yunlong Feng, co-investigator and assistant professor of mathematics and statistics, brings expertise in machine learning and data science to the project.
“Part of this work will involve comparing dental plaque images from healthy human patients against patients with periodontal disease,” Feng explains. “We will also analyze images of dental plaque from dogs, which host different microbial communities than people. This sort of comparison will help us identify patterns in biofilm structure — independent of bacterial species taxonomy — associated with gum health and disease. This understanding could open new pathways for improving periodontal disease treatments.”
The award is a Stephen I. Katz Early Stage Investigator Research Project Grant, a funding mechanism specifically designed to support early-career scientists looking to pursue a new line of research.
Valm concludes, “After landing here at UAlbany, I realized the tremendous research infrastructure and expertise that we have in RNA biology. Therefore, I have chosen to take full advantage of my situation in the RNA Institute to study the spatial patterning of gene expression in complex dental plaque communities. This work represents an exciting new direction for my lab and career.”
Alex Valm and Yunlong Feng will undertake this work alongside co-investigator Loreto Abusleme from the University of Chile.




